Medical Imaging
Description
The interactive visualization of large 3D medical data sets such as CT or
MRI scans is still a challenge. The problems mainly arise from the sheer size
of the data sets which have to be stored and processed somehow. Volume
visualization techniques, such as isosurface extraction, quickly encounter
the limits of current graphics hardware.
In order to be able to visualize (and process) these large data sets
efficiently, multiresolution techniques and parallelization are necessary.
However, usually these two paradigms don't match very well. Another problem
in the parallel context is load balancing, that is the problem of processors
getting idle at any time during visualization. We developed a parallel
multiresolution algorithm with dynamic load balancing which achieves an almost
optimal speedup on shared memory parallel computers.
Examples
If below pictures appear scrambled, please increase the width of your web
browser. Click on the pictures to see an enlarged version.



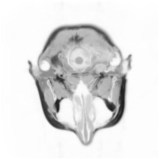
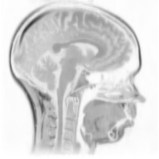
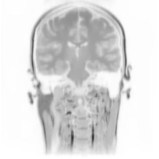
Above you see adaptive slices along various directions through a 3D MRI
scan of a human head (courtesy of MeVis, Bremen). Note that the slicing planes
can of course be oriented in any direction. The upper row contains the grids
corresponding to the grey scale images in the lower row. The adaptive method
requires much less triangles to represent the data with only small error in
comparison to a full triangulation (which would be a completely black grid).
The multiresolution algorithm allows an online adjustment of the error
threshold leading to finer or coarser images with higher or lower triangle
counts, respectively.

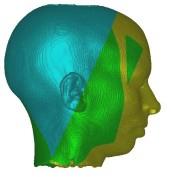
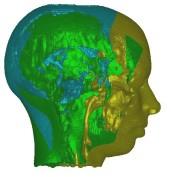


In these images you see isosurfaces of the same data set. Colors indicate
portions of the isosurface that have been extracted a the different processor
(3 in the upper row, 6 in the lower row). Note that the portions can change
between the images since the load balancing is dynamic and the workload
of the processors can change with time depending on the scheduler of the
operating system. For the rightmost picture of the upper row the front of
the head has been clipped away in order to show its interior.
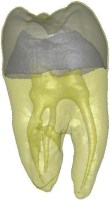
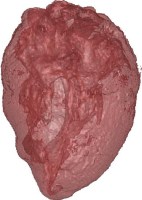
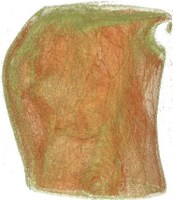
Above you see transparent isosurface renderings of a tooth, a sheep's heart and
a knee (data sets by B. Lorensen from the transfer function bake-off). The
isovalues of these images have been selected automatically based on average
isosurface normals.
References
-
T. Gerstner, Multiresolution extraction and rendering of transparent isosurfaces, Computers & Graphics, 26(2):219-228, 2002. (shortened version in "Data Visualization '01", D.S. Ebert, J.M. Favre and R. Peikert (eds.), pp. 35-44, Spinger, 2001).
-
T. Gerstner, R. Pajarola,
Topology preserving and controlled
topology simplifying multiresolution isosurface extraction, in
Proceedings IEEE Visualization 2000, pp. 259-266, IEEE Computer Society Press,
2000.
-
T. Gerstner, M. Rumpf,
Multiresolutional
parallel isosurface extraction based on tetrahedral bisection,
in Volume Graphics, M. Chen, A. Kaufman, and R. Yagel (eds.), Springer,
2000.
-
H. Pfister, W.E. Lorensen, C. Bajaj, G. Kindlmann, W.J. Schroeder, L.S.
Avila, K.M. Martin, Transfer Function Bake-Off, IEEE Computer
Graphics & Applications, 21(3), 2001.
Related Projects